Part 7. Global optimization: how to combine multiple data streams?
At LSCE we also have implemented a global Carbon Cycle Data Assimilation System (CCDAS), which uses many of the different data streams that we've already looked at, plus atmospheric CO2 concentration data from flask measurements around the globe. For this data stream we need to couple the ORCHIDEE model to an atmospheric transport model (LMDz) which transports the net surface CO2 fluxes in the atmosphere, in order to compare to the atmospheric concentration measurements at each station. Figure 7 shows a schematic of the overall CCDAS.
In the first version of the CCDAS we simultaneously assimilate (i.e. all data streams in the same optimization) FluxNet data (as described in Part 4), satellite-derived MODIS NDVI data and the atmospheric CO2 concentrations. The NDVI data are used to constrain the leaf phenology (leaf onset and senescence) at 15 sites per deciduous PFT across the globe.
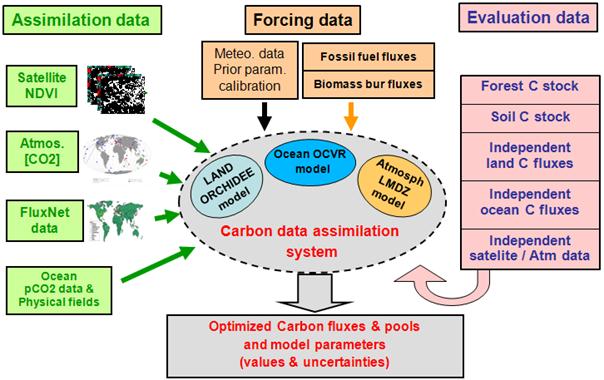
Figure 7. Schematic of the LSCE CCDAS
Figure 8 shows the results when optimizing just with atmospheric [CO2] data, then with both atmospheric [CO2] and MODIS NDVI data, and then with all three datastreams (including FluxNet).
Take a look and closely compare the figure and then answer the questions below.
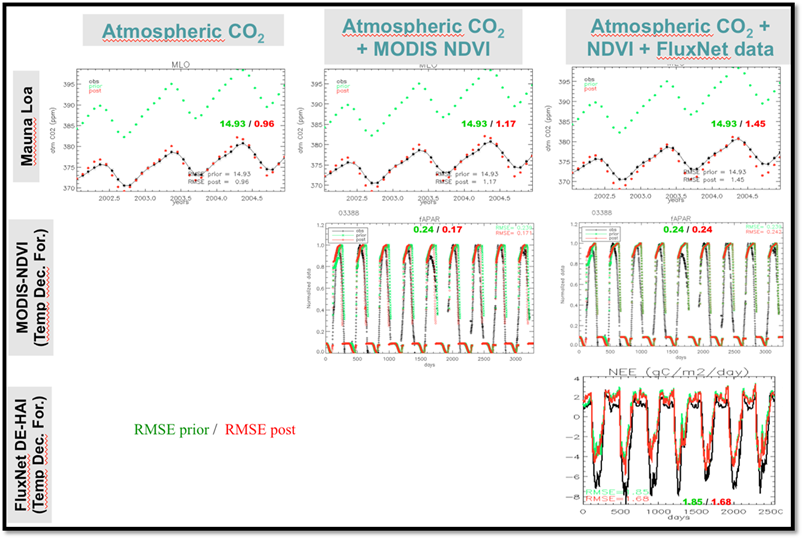
Figure 8. Results from different optimization using different data streams: First column with only atmospheric CO2 observation (70 sites); second column with atmospheric CO2 and NDVI from MODIS (taken for xx sites); Third column with atmospheric CO2, MODIS-NDVI and FluxNet Net Ecosystem Exchange and Latent Heat fluxes at around 70 sites. Each line display the fit to the data (prior and posterior) at only one station (Mauna Loa) for the atmospheric station, one site for NDVI (Deciduous broadleaf forest) and one site for the FluxNet NEE (Hainich in Germany).
- Why does adding data streams lower the reduction in RMSE for the atmospheric data?
- What could be the reasons explaining the poor fit to the NEE and NDVI data, when all data streams are optimized together?
- What are the difficulties in combining different observation data streams? What do you think are the crucial points to consider when set up the optimization with multiple data streams?
References
Reichstein, M., Falge, E., Baldocchi, D., Papale, D., Aubinet, A., Berbigier, P., Bernhofer, C., Buchmann, N., Gilmanov, T., Granier, A., Grünwald, T., Havránková, K., Ilvesniemi, H., Janous, D., Knohl, A., Laurila, T., Lohila, A., Loustau, D., Matteucci, G., Meyers, T., Miglietta, F., Ourcival, J.-M., Pumpanen, J., Rambal, S., Rotenberg, E., Sanz, M., Tenhunen, J., Seufert, G., Vaccari, F., Vesala, T., Yakir, D., and Valentini, R. (2005). On the separation of net ecosystem exchange into assimilation and ecosystem respiration: review and improved algorithm. Global Change Biol., 11, 1424-1439.







